| 2 |
Refresher |
| 2.1 |
Preliminary note |
This chapter reviews some key concepts from second year biochemistry. Feel free to skip it if you remember a thing or two from that distant past.
| 2.2 |
How enzymes work: active sites and catalytic mechanisms |
As with all proteins, the activity of enzymes depends on the precise arrangement and interaction of their amino acid residues and side chains. A straightforward example of this is chymotrypsin. Chymotrypsin is one of the major proteases in the human digestive tract, where its job is to knock down large protein molecules into small peptides that are then further processed by peptidases.
| 2.2.1 |
The “catalytic triad” in the active site of chymotrypsin |
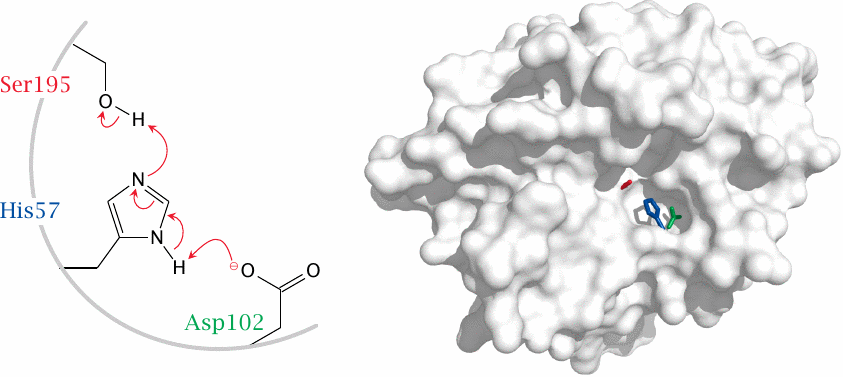
The active site of chymotrypsin contains aspartate 102, histidine 57, and serine 195. The aspartate and the histidine cooperate to deprotonate the hydroxyl group of the serine, which then attacks the substrate peptide bond (see next slide). Molecular structure rendered from 1afq.pdb.
The Asp-His-Ser motif is very common among proteases and esterases, so much so that it is often simply referred to as the catalytic triad. For example, the protease trypsin and several lipases that occur in human metabolism also have this motif and share the same mechanism of catalysis. Other enzymes, for example the proteasome, may contain glutamic acid instead of aspartic acid, or threonine instead of serine. These variants still contain the same functional groups and work the same way.
| 2.2.2 |
The catalytic mechanism of chymotrypsin |
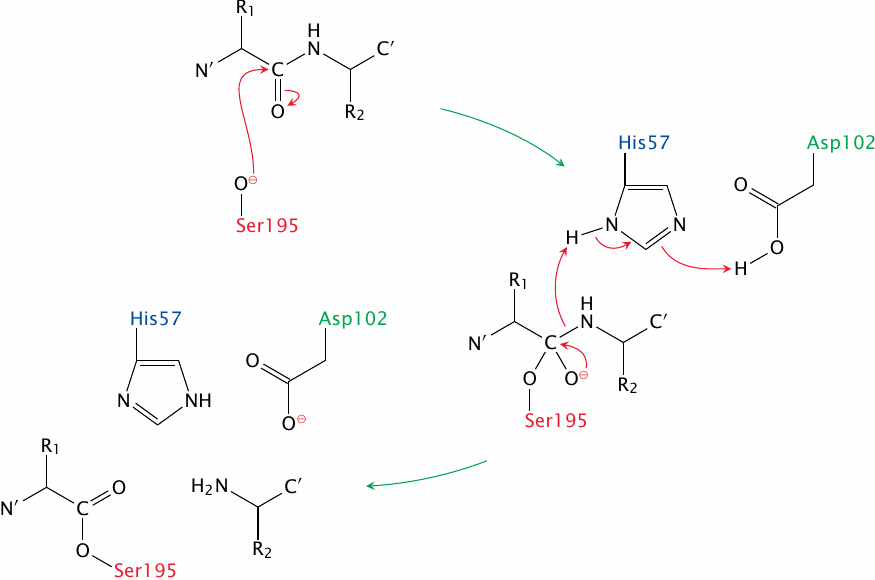
After it has been deprotonated by aspartate and histidine, the serine performs a nucleophilic attack on the carbonyl group of the substrate peptide bond. This produces a short-lived tetrahedral intermediate that gives way when the C-terminal peptide fragment leaves; the N-terminal fragment remains covalently attached to the serine. This state of affairs, which is shown as the final stage in this slide, is reached after the first half of the reaction.
In the second half reaction, which is not shown, the aspartate and histidine residues deprotonate a water molecule, and the hydroxide ion thus formed then bounces the N-terminal peptide fragment off the serine, again by nucleophilic attack on the carbonyl group. The bond undergoing cleavage at this stage is an ester, which yields more readily than the amide bond in the first stage.
| 2.2.3 |
Many enzymes require coenzymes |
With chymotrypsin, the enzyme molecule and its amino acid side chains supply all the necessary tools for catalysis. In contrast, many other enzyme molecules require coenzymes for their activity. For example, alanine aminotransferase, which transfers the α-amino group from alanine to α-ketoglutarate, contains the coenzyme pyridoxal phosphate within its active site. In the reaction, the coenzyme cooperates with a lysine reside that is part of the enzyme itself (see slide 12.2.1).
Most enzyme molecules have just one active site, or, in case they are multimeric, one active site per subunit. However, there are exceptions: Fatty acid synthase has as many as six different active sites on each subunit (see slide 10.5.2). Pyruvate dehydrogenase is a multienzyme complex that contains one active site on each subunit, but it combines three different types of subunits, each with a different coenzyme and catalytic function, into one functional assembly (see slide 5.2.2).
| 2.2.4 |
IUBMB classification of enzymes |
| Enzyme class | Catalyzed reactions |
| oxidoreductases | catalyze redox reactions, frequently involving one of the coenzymes NAD+, NADP+, or FAD |
| transferases | transfer functional groups between metabolites, e.g. a phosphate from ATP to a sugar hydroxyl group |
| hydrolases | catalyze hydrolysis reactions, such as those involved in the digestion of foodstuffs |
| lyases | perform elimination reactions that result in the formation of double bonds |
| isomerases | facilitate the interconversion of isomers |
| ligases | form new covalent bonds at the expense of ATP hydrolysis |
The IUBMB nomenclature divides all enzymes into six classes according to the reactions they catalyze. Within each of these main classes, there are subclasses and sub-sub classes, which reflect differences in substrate usage and mechanism of catalysis. The categories at all three hierarchical levels are assigned unique numbers, and each individual enzyme receives a number as well that is unique within its sub-sub class. An enzyme can therefore be unequivocally identified by a dot-separated identifier containing four numbers overall. This identifier is prefixed with the letters “EC” (for “Enzyme Commission”). Fittingly, the identifier EC 1.1.1.1 goes to the single most important enzyme in student lifestyle—namely, alcohol dehydrogenase, or, as IUBMB puts it, alcohol:NAD oxidoreductase.10 A list of all the enzyme activities recorded by the IUBMB classification is available at chem.qmul.ac.uk/iubmb.
The IUBMB scheme also assigns explicit names, which can be rather formidable, such as this one for the enzyme otherwise known as transketolase (see chapter 9): sedoheptulose-7-phosphate: d-glyceraldehyde-3-phosphate glycolaldehydetransferase. Reciting this name three times in quick succession will cure any stammer; however, we will forgo this benefit and use the traditional shorter names in this text.
| 2.3 |
Energetics of enzyme-catalyzed reactions |
With each enzymatic reaction, as with any other chemical reaction, energy comes in with these two questions: 1)will the reaction proceed at all in the desired direction, and 2)if it does, will it proceed at a sufficient rate?
The first question is decided by the free energy of the reaction, ΔG; a reaction will go forward if, and only if, the associated ΔG is negative. The second question depends on the activation energy, ΔG*, which forms a barrier between the initial state and the final state of the reactants. The very short-lived, energy-rich state at the top of this barrier is called the transition state. Enzymes can substantially lower the activation energy ΔG* and thus accelerate reactions, but they cannot change the overall free energy ΔG—and therefore, the direction or equilibrium—of the reaction.
| 2.3.1 |
A simile: the Walchensee–Kochelsee hydroelectric power system |
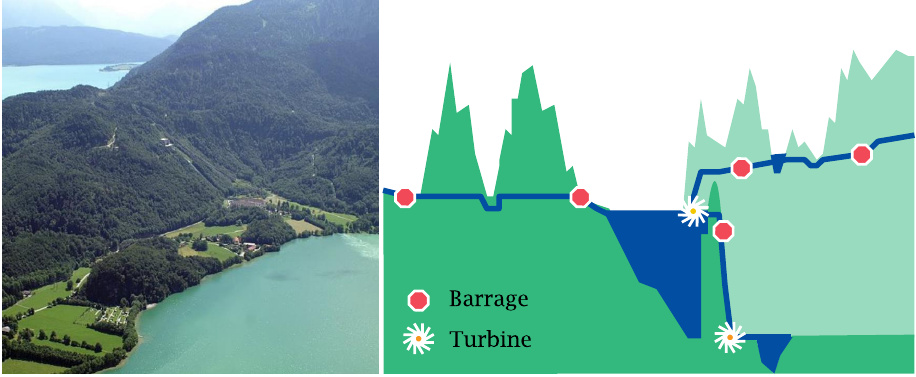
The different roles of ΔG and ΔG* in biochemical reactions can be illustrated with a simile. The slide shows two natural lakes in the German Alps. The Walchensee is situated 200 m above the Kochelsee. A conduit was dug across the barrier between these two lakes to make the water flow downhill and drive a hydroelectric turbine. Additional tunnels drain other lakes and rivers to enhance the supply of water to the Walchensee.
| 2.3.2 |
Analogies in the simile |
| Hydroelectric system | Metabolic pathway |
| altitude | energy |
| difference in altitude between lakes | energy difference between metabolites (ΔG) |
| height of ridge between lakes | ΔG* of uncatalyzed reaction |
| tunnels | enzymes |
| tunnel barrages | regulatory switches of enzymes |
An enzyme facilitates the interconversion of metabolites by creating an energetic “tunnel” across the energy barrier between them. Like a water conduit, an enzyme can facilitate the flow. However, it cannot change its direction, which will depend solely on the difference in altitude (ΔG).
In the hydro-electric system, tunnels can be opened or shuttered by barrages to accommodate variations in the amount of rainfall or in energy demand. Similarly, enzymes have regulatory switches that allow for adjustments in the flow rate through metabolic pathways in keeping with changing physiological needs.
| 2.3.3 |
Discrepancies in the simile |
| Hydroelectric system | Metabolic pathway |
| all tunnels work the same way | enzymes have different catalytic mechanisms |
| potential energy determined by one parameter: altitude | free energy of metabolites depends on two parameters: ΔH and ΔS |
| water always collects at the bottom | molecules partition between lower and higher energy levels |
Our simile illustrates some, but not all aspects of enzyme reactions. For example, all tunnels are alike; in contrast, each enzyme needs a specific “trick” or catalytic mechanism in order to accomplish the specific task at hand. Investigating the catalytic mechanisms of individual enzymes is an important and fascinating aspect of biochemistry.
Another difference concerns the energetic states. The energy difference between the two lakes is completely determined by their difference in altitude. However, the difference in free energy (ΔG) between two metabolites also depends on entropy, which is determined by their concentrations. Reactions are therefore subject to equilibrium; states with higher ΔH are less populated, but never totally unoccupied. The equilibrium is given by this relationship:
| \(\frac{n_1}{n_2}\) | = | \(e^{-\frac{\Delta G}{\text{R}T}}\) | (2.1) |
where n1 and n2 represent the numbers of molecules in the high and low energy states, respectively. (R is the gas constant, whereas T is the absolute temperature.)
Equation 2.1 applies to the occupancy of the initial and the final states of a reaction. It also applies to the distribution of molecules between the initial state and the transition state of a reaction. The tendency of molecules to spontaneously populate states of higher energy explains that chemical reactions will occur at all, even though the energy level of the transition state is always higher than those of the initial and final states. However, the higher the activation energy, the more rarefied the transition state will become. The number of molecules that can first climb the barrier and then hop down on the other side thus becomes smaller, and the reaction slower with increasing activation energy. Enzymes—and catalysts in general—create transition states that are lower in energy and therefore more populated than the uncatalyzed ones.
| 2.4 |
The role of ATP in enzyme-catalyzed reactions |
As we have seen, enzymes alone cannot drive endergonic reactions forward; however, an enzyme may couple an intrinsically endergonic reaction to an exergonic one, so as to make the overall reaction exergonic also. Most commonly, the auxiliary exergonic reaction consists in the hydrolysis of ATP to ADP or AMP. While this use of ATP pervades all of enzymology, it is important to understand that there is no equally general chemical mechanism of ATP utilization: each enzyme needs to find its own way of actually, chemically linking ATP hydrolysis to the reaction which it needs to drive.
| 2.4.1 |
The catalytic mechanism of glutamine synthetase |
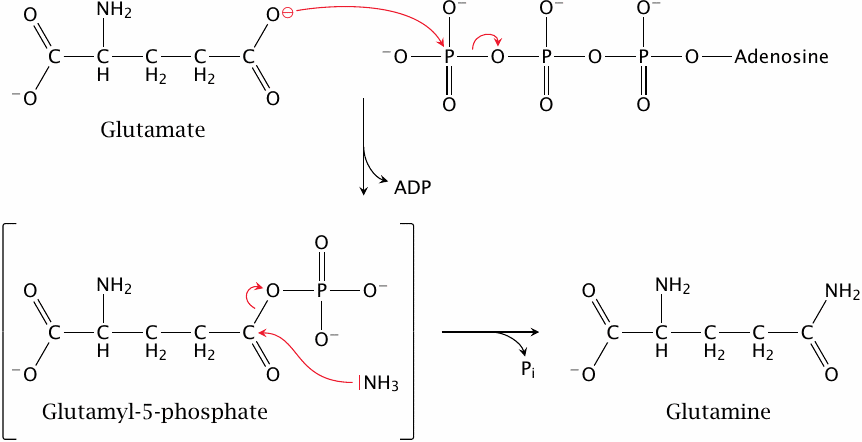
As an example, this slide shows how glutamine synthetase uses ATP to produce glutamine from glutamate and ammonia.
While the net turnover of ATP is hydrolysis, the ATP molecule isn’t hydrolyzed directly. Instead, the phosphate group is first transferred to the substrate to create an intermediate product, glutamyl-5-phosphate. In this mixed anhydride, the phosphate group makes a very good leaving group, which facilitates its subsequent substitution by ammonia. Therefore, the utilization of ATP is a central part of this enzyme’s catalytic mechanism. We will see some more examples of ATP usage in enzyme catalysis in the remainder of this notes.
| 2.5 |
Regulation of enzyme activity |
Just as a hydroelectric power station has to adjust to variations in water supply and demand for electricity, metabolic pathways and enzymes must adapt to changes in substrate availability and in demand for their products. The activities of enzymes are regulated at different levels. Activating gene expression will increase the abundance of an enzyme, whereas activation of protein breakdown will decrease it. In addition, there are mechanisms for reversibly activating or inactivating existing enzyme molecules, which enable swifter and potentially less wasteful adaptation. These reversible mechanisms are discussed in the following slides.
| 2.5.1 |
The phosphofructokinase reaction |
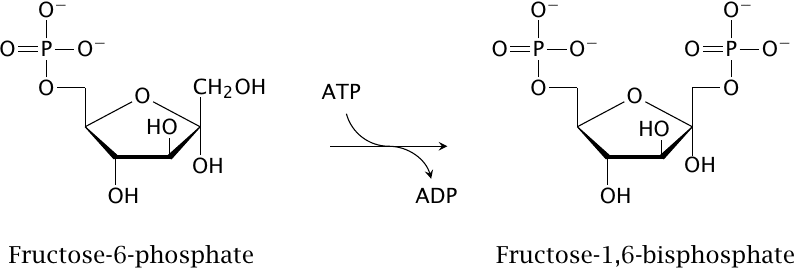
The enzyme phosphofructokinase transfers a phosphate group from ATP to fructose-6-phosphate, producing fructose-1,6-bisphosphate. This reaction occurs as an early step in the degradation of glucose. Since this pathway ultimately serves to regenerate ATP from ADP and phosphate, one might expect phosphofructokinase to be activated by ADP. However, it turns out that activation is mediated by AMP instead. The rationale for this preference is discussed in the next slide.
| 2.5.2 |
The adenylate kinase reaction equilibrates AMP, ADP and ATP |
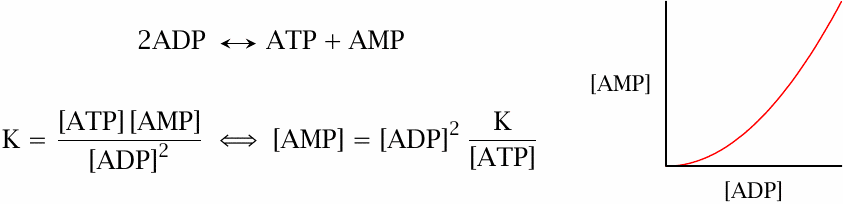
When ATP is consumed and ADP levels rise, some ATP can be regenerated by adenylate kinase, which turns two ADP molecules into one ATP and one AMP. According to the law of mass action, this also means that AMP levels rise quadratically with the level of ADP (assuming that changes to the level of ATP are small, which is usually the case). Its steeper rise makes AMP a better sensor of cellular energy demand than ADP itself, and it therefore makes sense that AMP, not ADP regulates the activity of phosphofructokinase.
| 2.5.3 |
Allosteric regulation of phosphofructokinase by AMP |
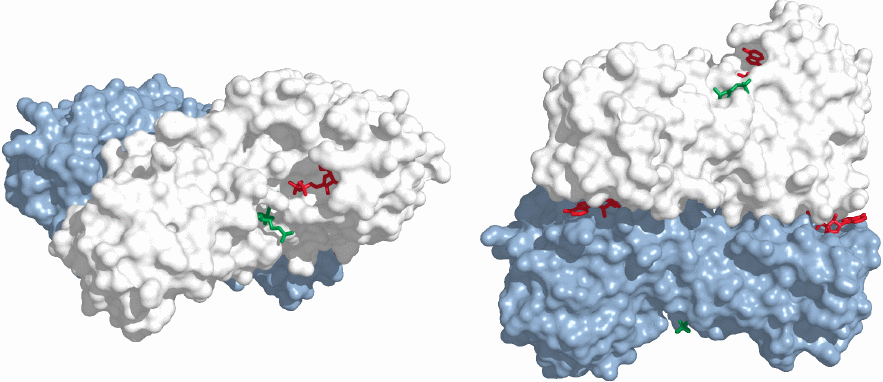
To accomplish the stimulation of phosphofructokinase, AMP interacts with the enzyme at an allosteric binding site, that is, a site that is distant and functionally distinct from the active site. In this structure of the dimeric enzyme (rendered from 1pfk.pdb), two molecules of ADP are bound for each enzyme subunit. The left panel shows an ADP molecule (red) bound in one of the active sites, which also contains the the other product of the reaction (fructose-1,6-bisphosphate, green). The side view in the right panel shows two more ADP molecules bound at the interface of the two enzyme subunits, within the allosteric binding sites that in the cell would bind AMP rather than ADP. The fourth ADP molecule in the second active site is hidden from view.
The adenine nucleotides bound at the two different sites assume entirely different roles. The ADP in the active site participates in the reaction. The AMP in the allosteric site does not; instead, its job is to change the conformation of the entire enzyme molecule. This conformational change will be transmitted through the body of the protein to the active site and increase the efficiency of catalysis there.
Allosteric regulation of enzymes is exceedingly common; it is not limited to nucleotides or any other particular class of metabolites. Allosteric effectors can be either stimulatory, as is AMP in this example, or inhibitory. As an example of the latter, ATP is not only a cosubstrate but also an allosteric inhibitor of phosphofructokinase.
Considering that the main purpose of the degradative pathway downstream of phosphofructokinase is regeneration of ATP, it makes sense to reduce the substrate flow through this pathway when ATP levels are high. The allosteric effect of ATP on phosphofructokinase is an example of feedback inhibition, that is, the inhibition of an early step in a pathway by that pathway’s main product. This is a very common principle in metabolic regulation.
| 2.5.4 |
How allosteric regulation works |
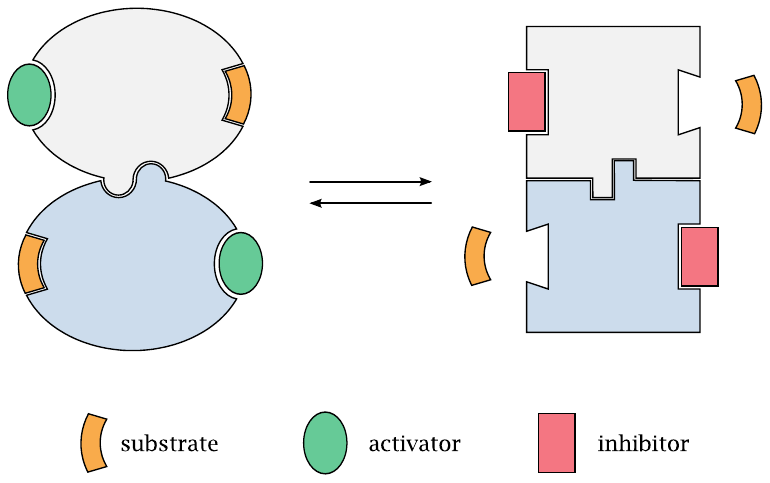
An allosterically regulated enzyme has two possible conformations that are in equilibrium with each other. Both the active site and the allosteric binding site change shape along with the molecule. An allosteric activator will bind selectively to the regulatory site in the shape that it assumes in the enzyme’s active conformation; the binding energy will shift the equilibrium towards this conformation. Conversely, an inhibitor will selectively bind and stabilize the enzyme’s inactive conformation.
As you can see from these considerations, activators and inhibitors may share the same regulatory site; with phosphofructokinase, this applies to ATP and AMP. Note, however, that human phosphofructokinase has an additional allosteric site that permits regulation by another effector (see slide 7.5.3).
| 2.5.5 |
Enzyme regulation by protein phosphorylation |
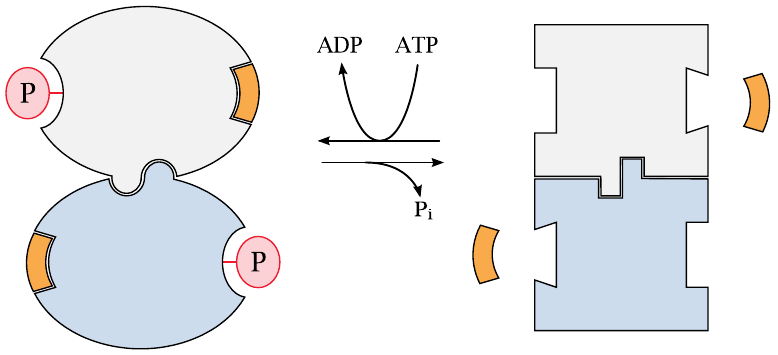
Another important means of regulating enzyme activity is through phosphorylation of the enzyme molecules. This is mediated by protein kinases, which transfer a phosphate group from ATP to specific amino acid side chains on the regulated enzymes.
When considering how protein phosphorylation works, it is best to think of the transferred phosphate group as an allosteric effector that happens to be covalently attached to the enzyme. Like proper allosteric regulators, the phosphate group imposes a conformational change that is transmitted to the active site through the body of the protein; and in both cases, it depends entirely on the enzyme in question whether it responds to the effector with an increase or a decrease in activity. For example, ATP allosterically inhibits phosphofructokinase, but it activates the functionally opposite enzyme fructose-1,6-bisphosphatase (see slide 7.5.3). Similarly, phosphorylation inhibits glycogen synthase but activates glycogen phosphorylase, which degrades glycogen (slide 8.4.1).
The major difference between allosteric regulation on the one hand and protein phosphorylation on the other is in the duration; an allosteric effector will dissociate as soon as its concentration drops, whereas phosphorylation will remain in effect until it is reversed by a specific protein phosphatase. Another, less obvious difference is that phosphorylation can apply to multiple sites in one protein. For example, in endothelial nitric oxide synthase (see slide 9.3.5), phosphorylation of alternate sites causes either activation or inhibition, respectively; this is managed by separate, site-specific protein kinases. In contrast, multiple and strictly alternate sites are not feasible with non-covalently binding allosteric effectors.
| 2.5.6 |
Oligomeric enzymes behave cooperatively |
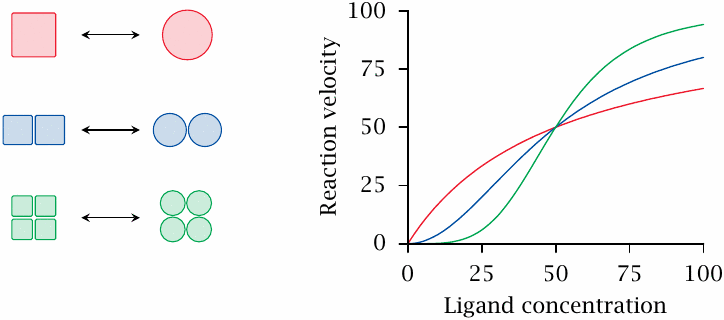
While allosteric control is in principle feasible with both monomeric and oligomeric enzyme molecules, almost all allosteric enzymes are indeed oligomeric proteins. Phosphofructokinase is a dimer; this is not uncommon, but often the number of subunits is considerably larger. Oligomeric enzymes usually respond cooperatively to effector binding, which means that all subunits change conformation simultaneously. This enables the enzymes to react more sensitively to small changes in effector concentration. Cooperative responses may be observed not only with allosteric effectors but also with substrates.
This slide illustrates theoretical dose-response curves for monomeric, dimeric and tetrameric enzymes. Each subunit is assumed to bind the ligand with the same affinity; the differences in curve shape arise from cooperativity alone. Note, however, that cooperativity may be partial, which means that oligomer subunits retain a degree of independence. Partial cooperativity results in experimental dose-response curves that are not as steep as theoretically possible.
| 2.5.7 |
Substrate cycles can amplify molecular regulation mechanisms |
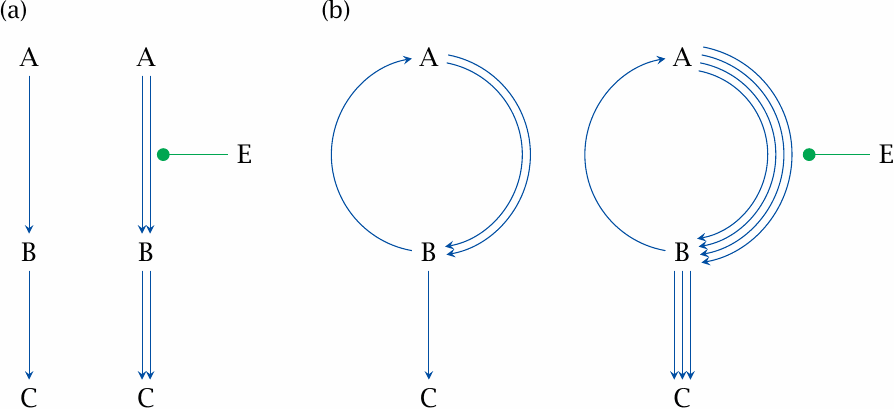
Cooperativity is one device for increasing the sensitivity of metabolic flux to regulation; substrate cycles are another. This is illustrated here for activation, but it applies similarly to inhibitory effectors as well. Let’s consider a simple, hypothetical pathway A⟶B⟶C (panel a). The rate is limited by the first reaction (A⟶B), so that the basal throughput of both reactions is the same; we may assume that this flow rate equals 1. If we apply an effector E that doubles the rate of the forward reaction A⟶B, the subsequent reaction B⟶C will be accelerated by the same factor, since we assumed the first reaction to be rate-limiting.
We can achieve the same basal throughput for B⟶C as in (a) using a substrate cycle between A and B, in which one enzyme converts A to B with a flow rate of 2, and a second enzyme converts B back to A with a rate of 1 (panel b). If we now add the same effector as in (a) and accordingly double the flow rate A⟶B, we obtain a flow rate of 4 from A to B. Diminished by the unchanged flow B⟶A, the resulting net flow rate for B⟶C becomes 3. The substrate cycle therefore amplifies the increase in metabolic flux in response to the same regulatory effect of E. This regulatory mechanism can be made even more effective by subjecting the step B⟶C to inhibition by E; such a pattern is observed for example with the substrate cycle formed by phosphofructokinase and fructose-1,6-bisphosphatase (slide 7.5.3).
Substrate cycles occur in several places in metabolism; we will see some examples in sections 6.10 and 7.5. In order to crank such a cycle, some energy must be expended—for example, the forward reaction may hydrolyze ATP, while the reverse reaction does not regenerate it. This energy expenditure becomes the net effect of the cycle, and for this reason substrate cycles are also referred to as futile cycles. The energy is simply dissipated as heat, which to a degree may be useful, particularly in warm-blooded animals; however, the throughput of such cycles must always be kept in check in order to avoid excessive energy wastage.
| 2.5.8 |
Regulation of enzyme molecule abundance |
- transcriptional induction
- accelerated mRNA degradation
- ubiquitin ligation, followed by proteolytic degradation
While all of the mechanisms discussed above reversibly modulate the activity of existing enzyme molecules, an enzyme’s activity may also be varied by changing its abundance in the cell. Firstly, the transcription of the gene encoding the enzyme can be turned on or off. This mechanism is employed by many hormones, for example thyroid hormones or cortisol and other steroid hormones. Similarly, the stability of the messenger RNA encoding the enzyme can be up- or downregulated by RNA-binding proteins and other mechanisms [2], with the corresponding effects on the abundance of the enzyme molecules.
Enzyme molecules can also be tagged with a small protein named ubiquitin, which marks the protein for proteolytic degradation within the proteasomes. Hormones may affect the activity of an enzyme through more than one of these mechanisms. For example, insulin increases the activity of glycogen synthase by way of transcriptional induction, increasing mRNA stability, and inhibition of protein phosphorylation.
In the following chapters, we will discuss the details of regulation only with some selected enzymes. Nevertheless, please keep in mind that virtually all enzyme molecules are subject to one or more regulatory mechanisms. The importance of these molecular control mechanisms in the regulation of metabolism as a whole cannot be overstated.